Marcello DiStasio, MD, PhD

![]()

Department of Pathology
Yale School of Medicine
Address:
300 George St. Room 353D
New Haven, CT, 06510
[map]
Neuroimmunology
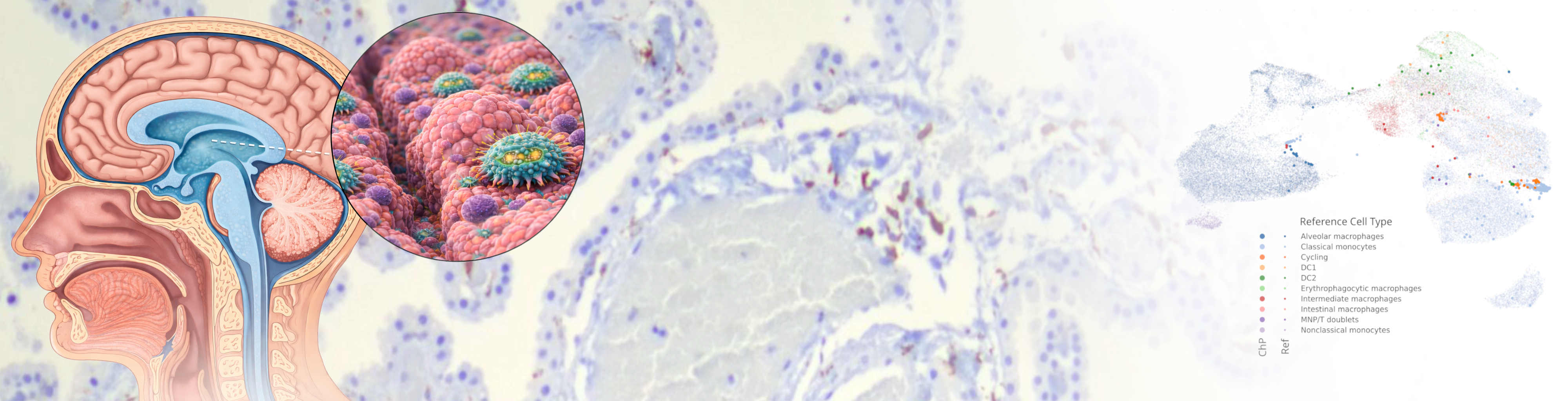
Our laboratory studies neuroimmunology with a focus on interactions at the borders of the central nervous system. We investigate how immune cells at CNS interfaces such as the choroid plexus, meninges, and perivascular spaces shape brain homeostasis and contribute to neurological disease.
Computational Biology
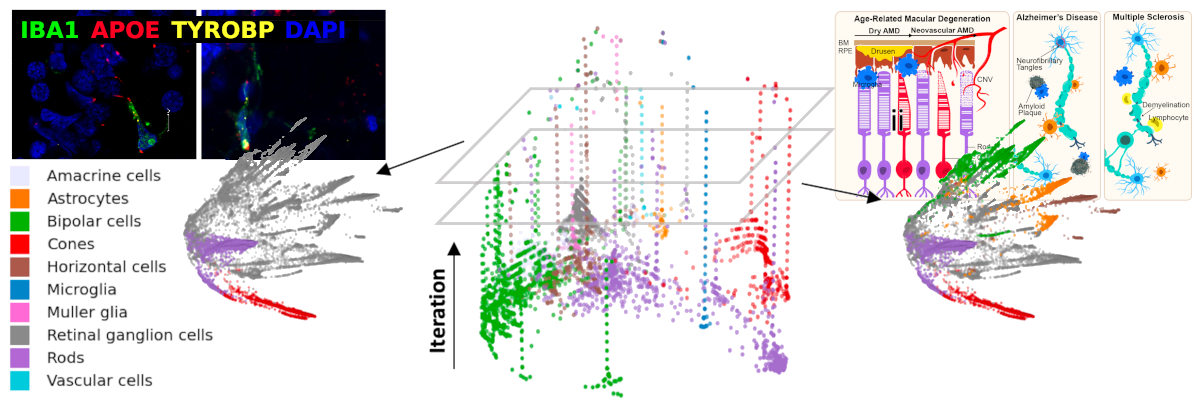
As part of the Yale Center for Neuroinflammation, our focus is on diseases of the central nervous system, including immune dysregulation, neurodegeneration, motor system diseases, and cancer. We develop new techniques in computational pathology in order to better understand and diagnose diseases. This involves the application of machine learning, image analysis, and statistics to histologic, genomic, clinical, and physiologic data. Our overall goal is to better characterize and classify the pathologic populations of central nervous system cells, and their interactions with their local microenvionment, to understand pathways implicated in disease and to uncover new therapeutic targets.
We develop software that analyzes genomic data in concert with histologic images taken from the kinds of slides produced in the routine clinical evaluation of tissue. By using statistical and machine learning techniques, these algorithms look for patterns in cell placement and morphology that correspond to the tissue genetic profiles. We believe this simultaneous genotypic and phenotypic characterization of tissue will provide a deeper understanding key microenvironments whose interactions advance our explanations and predictions of overall disease behavior and treatment response.
Digital Spatial Biology
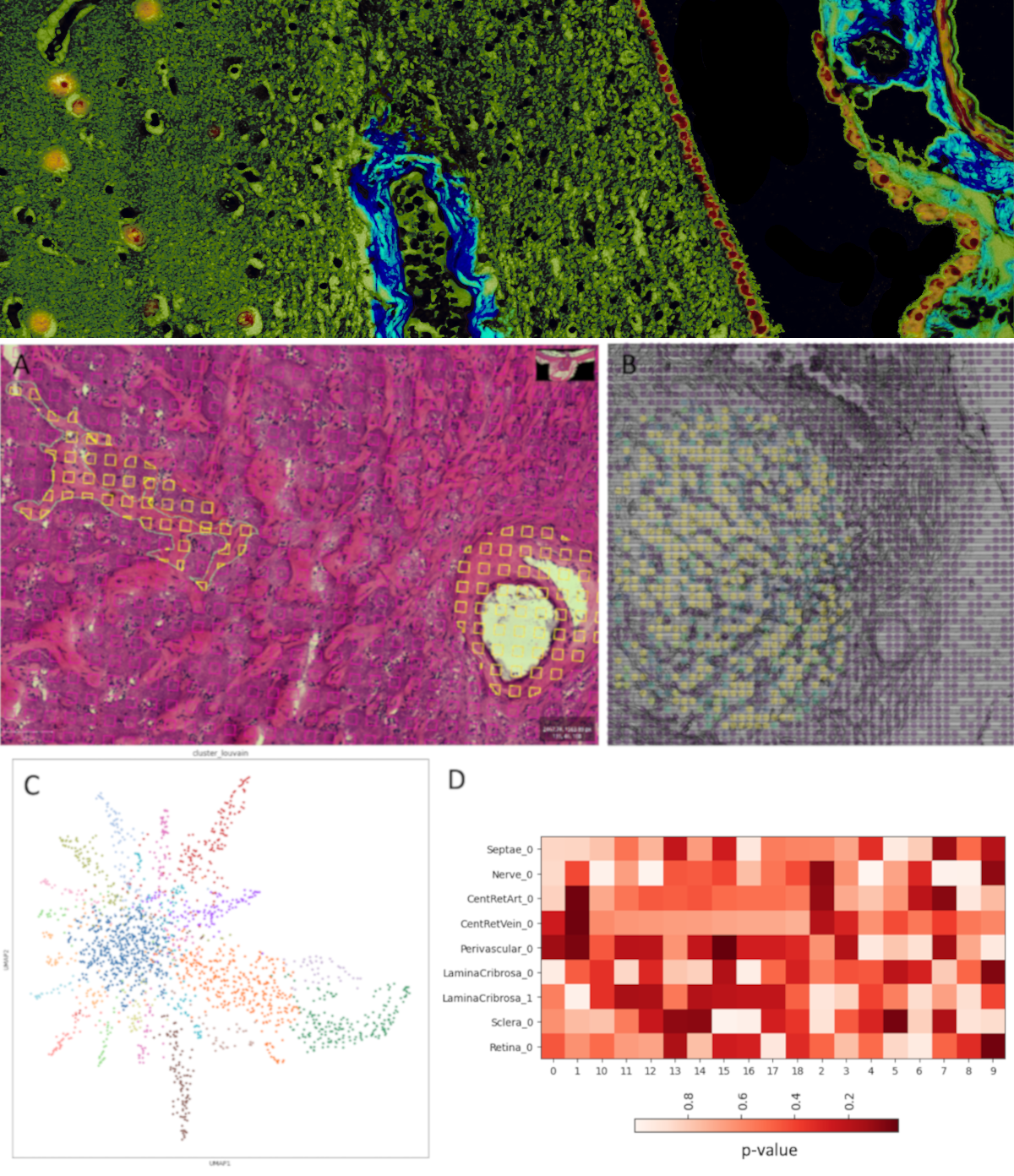
The recent development of technologies for highly multiplexed assays of nucleic acid sequences, chromatin state, and proteins while preserving information about their native locations in tissue (‘spatial -omics’ technologies) offers the tantalizing promise of new insights into biological processes (e.g. development) and the ways in which they become dysfunctional (i.e. pathology). This promise has re-emphasized the importance of tissue context to deliver true insights into disease biology.
The data generated by these technologies is a skeleton; the assays themselves provide the relative locations of the analyzed molecules (and thus proximities to each other). These molecules exist in and around cells which are situated in tissue architecture, creating the networks, physiological units, microenvironments, barriers, and organs which keep us alive. This context, which is, essentially, the traditional histologic description of health and disease, has been the subject of intense study since the 17th century, providing an immense body of knowledge of the changes in cellular identities, cytomorphology, and microenvironment that accompany disease initiation and progression.
Deployment of spatial ‘-omics’ technologies productively to understand molecular pathways implicated in disease requires application to tissue in which these structural elements can be identified and related to disease phenotype. To that end, we are creating tools to do two things: 1) Accurately align the highly multiplexed data generated by a spatial ‘-omics’ experiment with images of the tissue from which it was derived, and 2) Capture expert and/or automated identification of important locations and features in tissue and integrate them into the analysis of the molecular data in a meaningful way.
We develop tools which allows for localization and interpretation of the molecular data from highly multiplexed assays in the context of histologic images with annotations of pathologic features or morphologic characteristics such as lesions, breach of barriers, atrophy, or proximity to inflammation.
